⇒ April 2024
PhD position open in Fungal Genetics and Genomics
Understanding nitrate signaling in fungi
Fungi are one of the most active players in global nutrient cycling, and they can temporarily store nitrate in the soil by converting it to fungal biomass (known as nitrate assimilation process). Fungi therefore play a major role in preventing nitrogen losses from soil.
Despite the clear picture we already have on the regulatory genetic elements driving the nitrogen cycling process in the fungal cell, it is still not clear how the nitrate signal is perceived by the pathway transcriptional machinery. A new project financed by the Austrian Science Fund FWF sets out to study by molecular, genetic and biochemical approaches the NO3- signaling process that leads to activation of the pathway-specific transcription factor NirA. Structural biology work will include expression and crystallization of NirA wild-type and mutant variants and also cryo-EM experiments will be carried out to visualize structure-function relationships with interacting proteins. Genetic approaches are used to find suppressor mutations of regulatory elements and biochemical assays are planned to reveal functionally important intra- and intermolecular interactions. For the structural biology work, we intensely collaborate with specialized laboratories, among them Prof. Papageorgiou from the Turku Bioscience Centre in Finland where a research stay is planned during the PhD work.
If you are interested in the molecular genetics of how fungi act in this highly relevant ecological nitrogen cycling process, have a solid background in molecular biology and biochemistry and you would like to work in our group at a scientifically and socially stimulating working atmosphere, then please apply for this position.
Please send your application containing a motivation letter, your CV and a transcript of records per Email to joseph.strauss(at)boku.ac.at by latest End of May 2024. The position is scheduled for start by 1st of July 2024 and the salary is according to the FWF guidelines*.
⇒ June 2023
FWF project for Joseph Strauss
Nitrate signaling in fungi
Nitrate signaling in fungi
project number: P36690
decision board: 2023/06/05
keywords: nitrate, fungi, transcription factor, signal transduction
Publication from IMiG/BiMM
Genome analysis of Cephalotrichum gorgonifer and identifcation of the biosynthetic pathway for rasfonin, an inhibitor of KRAS dependent cancer
Background
Fungi are important sources for bioactive compounds that find their applications in many important sectors like in the pharma-, food- or agricultural industries. In an environmental monitoring project for fungi involved in soil nitrogen cycling we also isolated Cephalotrichum gorgonifer (strain NG_p51). In the course of strain characterisation work we found that this strain is able to naturally produce high amounts of rasfonin, a polyketide inducing autophagy, apoptosis, necroptosis in human cell lines and showing anti-tumor activity in KRAS-dependent cancer cells.
⇒ September 2022
Publication from Strauss group
The KdmB-EcoA-RpdA-SntB chromatin complex binds regulatory genes and coordinates fungal development with mycotoxin synthesis
Abstract
Chromatin complexes control a vast number of epigenetic developmental processes. Filamentous fungi present an important clade of microbes with poor understanding of underlying epigenetic mechanisms. Here, we describe a chromatin binding complex in the fungus Aspergillus nidulans composing of a H3K4 histone demethylase KdmB, a cohesin acetyltransferase (EcoA), a histone deacetylase (RpdA) and a histone reader/E3 ligase protein (SntB). In vitro and in vivo evidence demonstrate that this KERS complex is assembled from the EcoA-KdmB and SntB-RpdA heterodimers. KdmB and SntB play opposing roles in regulating the cellular levels and stability of EcoA, as KdmB prevents SntB-mediated degradation of EcoA. The KERS complex is recruited to transcription initiation start sites at active core promoters exerting promoter-specific transcriptional effects. Interestingly, deletion of any one of the KERS subunits results in a common negative effect on morphogenesis and production of secondary metabolites, molecules important for niche securement in filamentous fungi. Consequently, the entire mycotoxin sterigmatocystin gene cluster is downregulated and asexual development is reduced in the four KERS mutants. The elucidation of the recruitment of epigenetic regulators to chromatin via the KERS complex provides the first mechanistic, chromatin-based understanding of how development is connected with small molecule synthesis in fungi.
https://doi.org/10.1093/nar/gkac744
Publication from Strauss group
"How to Completely Squeeze a Fungus—Advanced Genome Mining Tools for Novel Bioactive Substances"
Abstract
Fungal species have the capability of producing an overwhelming diversity of bioactive substances that can have beneficial but also detrimental effects on human health. These so-called secondary metabolites naturally serve as antimicrobial “weapon systems”, signaling molecules or developmental effectors for fungi and hence are produced only under very specific environmental conditions or stages in their life cycle. However, as these complex conditions are difficult or even impossible to mimic in laboratory settings, only a small fraction of the true chemical diversity of fungi is known so far. This also implies that a large space for potentially new pharmaceuticals remains unexplored. We here present an overview on current developments in advanced methods that can be used to explore this chemical space. We focus on genetic and genomic methods, how to detect genes that harbor the blueprints for the production of these compounds (i.e., biosynthetic gene clusters, BGCs), and ways to activate these silent chromosomal regions. We provide an in-depth view of the chromatin-level regulation of BGCs and of the potential to use the CRISPR/Cas technology as an activation tool.
https://doi.org/10.3390/pharmaceutics14091837
Publication from Strauss group
"RimO (SrrB) is required for carbon starvation signaling and production of secondary metabolites in Aspergillus nidulans"
Abstract
Depending on the prevailing environmental, developmental and nutritional conditions, fungi activate biosynthetic gene clusters (BGCs) to produce condition-specific secondary metabolites (SMs). For activation, global chromatin-based de-repression must be integrated with pathway-specific induction signals. Here we describe a new global regulator needed to activate starvation-induced SMs. In our transcriptome dataset, we found locus AN7572 strongly transcribed solely under conditions of starvation-induced SM production. The predicted AN7572 protein is most similar to the stress and nutritional regulator Rim15 of Saccharomyces cerevisiae, and to STK-12 of Neurospora crassa. Based on this similarity and on stress and nutritional response phenotypes of A. nidulans knock-out and overexpression strains, AN7572 is designated rimO. In relation to SM production, we found that RimO is required for the activation of starvation-induced BGCs, including the sterigmatocystin (ST) gene cluster. Here, RimO regulates the pathway-specific transcription factor AflR both at the transcriptional and post-translational level. At the transcriptional level, RimO mediates aflR induction following carbon starvation and at the post-translational level, RimO is required for nuclear accumulation of the AflR protein. Genome-wide transcriptional profiling showed that cells lacking rimO fail to adapt to carbon starvation that, in the wild type, leads to down-regulation of genes involved in basic metabolism, membrane biogenesis and growth. Consistently, strains overexpressing rimO are more resistant to oxidative and osmotic stress, largely insensitive to glucose repression and strongly overproduce several SMs. Our data indicate that RimO is a positive regulator within the SM and stress response network, but this requires nutrient depletion that triggers both, rimO gene transcription and activation of the RimO protein.
https://doi.org/10.1016/j.fgb.2022.103726
⇒ August 2022
Publication from Strauss group
"Polaramycin B, and not physical interaction, is the signal that rewires fungal metabolism in the Streptomyces–Aspergillus interaction"
Abstract
Co-culturing the bacterium Streptomyces rapamycinicus and the ascomycete Aspergillus nidulans has previously been shown to trigger the production of orsellinic acid (ORS) and its derivates in the fungal cells. Based on these studies it was assumed that direct physical contact is a prerequisite for the metabolic reaction that involves a fungal amino acid starvation response and activating chromatin modifications at the biosynthetic gene cluster (BGC). Here we show that not physical contact, but a guanidine containing macrolide, named polaramycin B, triggers the response. The substance is produced constitutively by the bacterium and above a certain concentration, provokes the production of ORS. In addition, several other secondary metabolites were induced by polaramycin B. Our genome-wide transcriptome analysis showed that polaramycin B treatment causes downregulation of fungal genes necessary for membrane stability, general metabolism and growth. A compensatory genetic response can be observed in the fungus that included upregulation of BGCs and genes necessary for ribosome biogenesis, translation and membrane stability. Our work discovered a novel chemical communication, in which the antifungal bacterial metabolite polaramycin B leads to the production of antibacterial defence chemicals and to the upregulation of genes necessary to compensate for the cellular damage caused by polaramycin B.
https://doi.org/10.1111/1462-2920.16118
⇒ October 2021
BiMM paper on a new bioactive molecule from a new fungus
"Luteapyrone, a Novel ƴ-Pyrone Isolated from the Filamentous Fungus Metapochonia lutea"
Abstract
In the process of screening for new bioactive microbial metabolites we found a novel ƴ-pyrone derivative for which we propose the trivial name luteapyrone, in a recently described microscopic filamentous fungus, Metapochonia lutea BiMM-F96/DF4. The compound was isolated from the culture extract of the fungus grown on modified yeast extract sucrose medium by means of flash chromatography followed by preparative HPLC. The chemical structure was elucidated by NMR and LC-MS. The new compound was found to be non-cytotoxic against three mammalian cell lines (HEK 263, KB-3.1 and Caco-2). Similarly, no antimicrobial activity was observed in tested microorganisms (gram positive and negative bacteria, yeast and fungi).
» https://doi.org/10.3390/molecules26216589
⇒ July 2021
BIMM Paper on new fungi and their metabolites
"Polyphasic Approach Utilized for the Identification of Two New Toxigenic Members of Penicillium Section Exilicaulis, P. krskae and P. silybi spp. nov."
Abstract
Two new species, Penicillium krskae (isolated from the air as a lab contaminant in Tulln (Austria, EU)) and Penicillium silybi (isolated as an endophyte from asymptomatic milk thistle (Silybum marianum) stems from Josephine County (Oregon, USA)) are described. The new taxa are well supported by phenotypic (especially conidial ornamentation under SEM, production of red exudate and red pigments), physiological (growth at 37 °C, response to cycloheximide and CREA), chemotaxonomic (production of specific extrolites), and multilocus phylogenetic analysis using RNA-polymerase II second largest subunit (RPB2), partial tubulin (benA), and calmodulin (CaM). Both new taxa are resolved within the section Exilicaulis in series Restricta and show phylogenetic affiliation to P. restrictum sensu stricto. They produce a large spectrum of toxic anthraquinoid pigments, namely, monomeric anthraquinones related to emodic and chloremodic acids and other interesting bioactive extrolites (i.e., endocrocin, paxilline, pestalotin, and 7-hydroxypestalotin). Of note, two bianthraquinones (i.e., skyrin and oxyskyrin) were detected in a culture extract of P. silybi. Two new chloroemodic acid derivatives (2-chloro-isorhodoptilometrin and 2-chloro-desmethyldermoquinone) isolated from the exudate of P. krskae ex-type culture were analyzed by nuclear magnetic resonance (NMR) and liquid chromatography–mass spectrometry (LC–MS).
» https://doi.org/10.3390/jof7070557
"Molecular systematics of Keratinophyton: the inclusion of species formerly referred to Chrysosporium and description of four new species"
Abstract
Four new Keratinophyton species (Ascomycota, Pezizomycotina, Onygenales), K. gollerae, K. lemmensii, K. straussii, and K. wagneri, isolated from soil samples originating from Europe (Austria, Italy, and Slovakia) are described and illustrated. The new taxa are well supported by phylogenetic analysis of the internal transcribed spacer region (ITS) region, the combined data analysis of ITS and the nuclear large subunit (LSU) rDNA, and their phenotype. Based on ITS phylogeny, within the Keratinophyton clade, K. lemmensii is clustered with K. durum, K. hubeiense, K. submersum, and K. siglerae, while K. gollerae, K. straussii and K. wagneri are resolved in a separate terminal cluster. All four new species can be well distinguished from other species in the genus based on phenotype characteristics alone. Ten new combinations are proposed for Chrysosporium species which are resolved in the monophyletic Keratinophyton clade. A new key to the recognized species is provided herein.
» https://doi.org/10.1186/s43008-021-00070-2
"Biosynthesis of Fusapyrone Depends on the H3K9 Methyltransferase, FmKmt1, in Fusarium mangiferae"
The phytopathogenic fungus Fusarium mangiferae belongs to the Fusarium fujikuroi species complex (FFSC). Members of this group cause a wide spectrum of devastating diseases on diverse agricultural crops. F. mangiferae is the causal agent of the mango malformation disease (MMD) and as such detrimental for agriculture in the southern hemisphere. During plant infection, the fungus produces a plethora of bioactive secondary metabolites (SMs), which most often lead to severe adverse defects on plants health. Changes in chromatin structure achieved by posttranslational modifications (PTM) of histones play a key role in regulation of fungal SM biosynthesis. Posttranslational tri-methylation of histone 3 lysine 9 (H3K9me3) is considered a hallmark of heterochromatin and established by the SET-domain protein Kmt1. Here, we show that FmKmt1 is involved in H3K9me3 in F. mangiferae. Loss of FmKmt1 only slightly though significantly affected fungal hyphal growth and stress response and is required for wild type-like conidiation. While FmKmt1 is largely dispensable for the biosynthesis of most known SMs, removal of FmKMT1 resulted in an almost complete loss of fusapyrone and deoxyfusapyrone, γ-pyrones previously only known from Fusarium semitectum. Here, we identified the polyketide synthase (PKS) FmPKS40 to be involved in fusapyrone biosynthesis, delineate putative cluster borders by co-expression studies and provide insights into its regulation.
» https://doi.org/10.3389/ffunb.2021.671796
"Development and Validation of a Simple Bioaerosol Collection Filter System Using a Conventional Vacuum Cleaner for Sampling"
Abstract
Although numerous bioaerosol samplers for counting and identifying airborne microorganisms are available, the considerably high purchase and maintenance costs for the sampler often prevent broad monitoring campaigns for occupational or environmental surveillance of bioaerosols. We present here a newly developed simple adapter and filter system (TOP filter system) designed to collect bioaerosol particles from a defined air volume using conventional vacuum cleaners as air pumps. We characterized the physical properties of the system using air flow measurements and validated the biological performance. The culture-based detection capacities for airborne fungal species were compared to a standard impaction sampler (MAS-100 NT) under controlled conditions in a bioaerosol chamber (using Trichoderma spores as the test organism) as well as in the field. In the chamber, an overall equivalent detection capacity between all tested filters was recorded, although a significant underrepresentation of the TOP filter system for Trichoderma spores were seen in comparison to the MAS-100 NT. In a comparative field study (n = 345), the system showed similar biological sampling efficiencies compared to the MAS-100 NT impactor, only the diversity of identified fungal communities was slightly lower on the filters. Thus, the system is suitable for large-scale environmental sampling operations where many samples have to be taken in parallel at a given time at distant locations. This system would allow endeavors such as antibiotics resistance monitoring or hygiene surveys in agricultural or occupational settings.
» https://doi.org/10.1007/s41810-021-00110-9
⇒ June 2021
"The H4K20 methyltransferase Kmt5 is involved in secondary metabolism and stress response in phytopathogenic Fusarium species"
Highlights
- Kmt5 is solely responsible for H4K20me3 in F. fujikuroi and F. graminearum.
- Imbalanced H4K20me3 levels affect hyphal growth in F. graminearum.
- FgKMT5 deletion strains are hypovirulent on wheat in F. graminearum.
- H4K20me3 is required for wild type-like secondary metabolism in both fusaria.
- Mutants lacking Kmt5 are more tolerant to oxidative and osmotic stress.
» https://doi.org/10.1016/j.fgb.2021.103602
⇒ May 2021
"Chromatin Regulation and Epigenetics" - a special issue for "Fungal Genetics and Biology"
David Canovas and Joseph Strauss edited a special issue for "Fungal Genetics and Biology" with the title "Chromatin Regulation and Epigenetics"
https://www.sciencedirect.com/journal/fungal-genetics-and-biology/special-issue/10C73J2QZHJ
The issue and an extensive Editorial written by David and Joseph providing an overarching view on the published articles is out now.
https://www.sciencedirect.com/science/article/pii/S1087184521000530?via%3Dihub
⇒ March 2021
Publication with Contribution from Gergely Molnár
"AGC kinases and MAB4/MEL proteins maintain PIN polarity by limiting lateral diffusion in plant cells"
Highlights:
- MAB4/MEL proteins are recruited to the plasma membrane by PINs
- PINs, MAB4/MELs, and AGC kinases directly interact in a multiprotein complex
- PIN phosphorylation and MAB4/MEL recruitment form a positive feedback loop
- MAB4/MELs and AGC kinases maintain PIN polarity by limiting PIN lateral diffusion
» https://doi.org/10.1016/j.cub.2021.02.028
⇒ January 2021
Schimmelpilz auf dem Weg zum Lebensretter
Immer mehr Bakterien werden gegen die bekannten Antibiotika resistent und damit nicht mehr behandelbar. Das BIMM-Team in Tulln forscht mit Robotertechnik an Schimmelpilzen, die das ändern sollen. Sie könnten die Antibiotika der Zukunft werden – unter anderem.
» https://noe.orf.at/stories/3084889/
» BIMM
BiMM cooperation paper with Italian Fusarium group on the mycotoxin repressing function of thymol
"Naturally Occurring Phenols Modulate Vegetative Growth and Deoxynivalenol Biosynthesis in Fusarium graminearum"
This paper reports a couple of natural substances from plants that repress the production of the main Fusarium mycotoxin DON but does not inhibit growth of the fungus. Application perspectives are open and we also will try to understand the underlying mechanisms.
Abstract
To assess the in vitro activity of five naturally occurring phenolic compounds (ferulic acid, apocynin, magnolol, honokiol, and thymol) on mycelial growth and type B trichothecene mycotoxin accumulation by Fusarium graminearum, three complementary approaches were adopted. First, a high-throughput photometric continuous reading array allowed a parallel quantification of F. graminearum hyphal growth and reporter TRI5 gene expression directly on solid medium. Second, RT-qPCR confirmed the regulation of TRI5 expression by the tested compounds. Third, liquid chromatography–tandem mass spectrometry analysis allowed quantification of deoxynivalenol (DON) and its acetylated forms released upon treatment with the phenolic compounds. Altogether, the results confirmed the activity of thymol and an equimolar mixture of thymol–magnolol at 0.5 mM, respectively, in inhibiting DON production without affecting vegetative growth. The medium pH buffering capacity after 72–96 h of incubation is proposed as a further element to highlight compounds displaying trichothecene inhibitory capacity with no significant fungicidal effect.
» https://doi.org/10.1021/acsomega.0c04260
» https://www.bimm-research.at/
⇒ November 2020
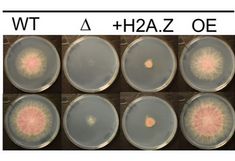
Novel genetic link between histone variant H2A.Z and chromatin remodelling found in Fusarium wheat pathogen
The collaborative paper with the Pont group from INRA-Bordeaux published in the October issue of PLOS-Genetics describes the H2A.Z histone variant in Fusarium graminearum, one of the most relevant fungal wheat pathogens worldwide (https://journals.plos.org/plosgenetics/article/authors?id=10.1371/journal.pgen.1009125).
H2A.Z is essential there, but it was possible to select strains that survive despite having a H2A.Z deletion.
WGS of several revertants revealed a number of possible compensatory mutations, all of them affected in components of the chromatin remodelling machinery. Hence, a new molecular link between H2A.Z and chromatin remodeller function is popping up.
⇒ October 2020
"Receptor kinase module targets PIN-dependent auxin transport during canalization"
Spontaneously arising channels that transport the phytohormone auxin provide positional cues for selforganizing aspects of plant development such as flexible vasculature regeneration or its patterning during leaf venation. The auxin canalization hypothesis proposes a feedback between auxin signaling and transport as the underlying mechanism, but molecular players await discovery. We identified part of the machinery that routes auxin transport. The auxin-regulated receptor CAMEL (Canalization-related Auxin-regulated Malectin-type RLK) together with CANAR (Canalization-related Receptor-like kinase) interact with and phosphorylate PIN auxin transporters. camel and canar mutants are impaired in PIN1 subcellular trafficking and auxin-mediated PIN polarization, which macroscopically manifests as defects in leaf venation and vasculature regeneration after wounding. The CAMEL-CANAR receptor complex is part of the auxin feedback that coordinates polarization of individual cells during auxin canalization.
» https://science.sciencemag.org/content/370/6516/550.full
"A novel fungal gene regulation system based on inducible VPR-dCas9 and nucleosome map-guided sgRNA positioning"
Abstract
Programmable transcriptional regulation is a powerful tool to study gene functions. Current methods to selectively regulate target genes are mainly based on promoter exchange or on overexpressing transcriptional activators. To expand the discovery toolbox, we designed a dCas9-based RNA-guided synthetic transcription activation system for Aspergillus nidulans that uses enzymatically disabled “dead” Cas9 fused to three consecutive activation domains (VPR-dCas9). The dCas9-encoding gene is under the control of an estrogen-responsive promoter to allow induction timing and to avoid possible negative effects by strong constitutive expression of the highly active VPR domains. Especially in silent genomic regions, facultative heterochromatin and strictly positioned nucleosomes can constitute a relevant obstacle to the transcriptional machinery. To avoid this negative impact and to facilitate optimal positioning of RNA-guided VPR-dCas9 to targeted promoters, we have created a genome-wide nucleosome map from actively growing cells and stationary cultures to identify the cognate nucleosome-free regions (NFRs). Based on these maps, different single-guide RNAs (sgRNAs) were designed and tested for their targeting and activation potential. Our results demonstrate that the system can be used to regulate several genes in parallel and, depending on the VPR-dCas9 positioning, expression can be pushed to very high levels. We have used the system to turn on individual genes within two different biosynthetic gene clusters (BGCs) which are silent under normal growth conditions. This method also opens opportunities to stepwise activate individual genes in a cluster to decipher the correlated biosynthetic pathway.
» https://doi.org/10.1007/s00253-020-10900-9
⇒ May 2020
Publication from Gergely Molnar
The lipid code-dependent phosphoswitch PDK1–D6PK activates PIN-mediated auxin efflux in Arabidopsis
Abstract
Directional intercellular transport of the phytohormone auxin mediated by PIN-FORMED (PIN) efflux carriers has essential roles in both coordinating patterning processes and integrating multiple external cues by rapidly redirecting auxin fluxes. PIN activity is therefore regulated by multiple internal and external cues, for which the underlying molecular mechanisms are not fully elucidated. Here, we demonstrate that 3′-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE1 (PDK1), which is conserved in plants and mammals, functions as a molecular hub that perceives upstream lipid signalling and modulates downstream substrate activity through phosphorylation. Using genetic analysis, we show that the loss-of-function Arabidopsis pdk1.1 pdk1.2 mutant exhibits a plethora of abnormalities in organogenesis and growth due to defective polar auxin transport. Further cellular and biochemical analyses reveal that PDK1 phosphorylates D6 protein kinase, a well-known upstream activator of PIN proteins. We uncover a lipid-dependent phosphorylation cascade that connects membrane-composition-based cellular signalling with plant growth and patterning by regulating morphogenetic auxin fluxes.
» https://doi.org/10.1038/s41477-020-0648-9
⇒ April 2020
"High Throughput Screening for New Fungal Polyester Hydrolyzing Enzymes"
here is a strong need for novel and more efficient polyester hydrolyzing enzymes in order to enable the development of more environmentally friendly plastics recycling processes allowing the closure of the carbon cycle. In this work, a high throughput system on microplate scale was used to screen a high number of fungi for their ability to produce polyester-hydrolyzing enzymes. For induction of responsible enzymes, the fungi were cultivated in presence of aliphatic and aromatic polyesters [poly(1,4-butylene adipate co terephthalate) (PBAT), poly(lactic acid) (PLA) and poly(1,4-butylene succinate) (PBS)], and the esterase activity in the culture supernatants was compared to the culture supernatants of fungi grown without polymers. The results indicate that the esterase activity of the culture supernatants was induced in about 10% of the tested fungi when grown with polyesters in the medium, as indicated by increased activity (to >50 mU/mL) toward the small model substrate para-nitrophenylbutyrate (pNPB). Incubation of these 50 active culture supernatants with different polyesters (PBAT, PLA, PBS) led to hydrolysis of at least one of the polymers according to liquid chromatography-based quantification of the hydrolysis products terephthalic acid, lactic acid and succinic acid, respectively. Interestingly, the specificities for the investigated polyesters varied among the supernatants of the different fungi.
» https://doi.org/10.3389/fmicb.2020.00554
» https://www.bimm-research.at/
⇒ January 2020
Paper about fungal development connected to nitric oxide
"Nitric oxide homeostasis is required for light-dependent regulation of conidiation in Aspergillus"
Abstract
Nitric oxide (NO) can be biologically synthesized from nitrite or from arginine. Although NO is involved as a signal in many biological processes in bacteria, plants, and mammals, still little is known about the role of NO in fungi. Here we show that NO levels are regulated by light as an environmental signal in Aspergillus nidulans. The flavohaemoglobin-encoding fhbB gene involved in NO oxidation to nitrate, and the arginine-regulated arginase encoded by agaA, which controls the intracellular concentration of arginine, are both up-regulated by light. The phytochrome fphA is required for the light-dependent induction of fhbB and agaA, while the white-collar gene lreA acts as a repressor when arginine is present in the media. The intracellular arginine pools increase upon induction of both developmental programs (conidiation and sexual development), and the increase is higher under conditions promoting sexual development. The presence of low concentrations of arginine does not affect the light-dependent regulation of conidiation, but high concentrations of arginine overrun the light signal. Deletion of fhbB results in the partial loss of the light regulation of conidiation on arginine and on nitrate media, while deletion of fhbA only affects the light regulation of conidiation on nitrate media. Our working model considers a cross-talk between environmental cues and intracellular signals to regulate fungal reproduction.
» https://doi.org/10.1016/j.fgb.2020.103337
Publication from Gergely Molnar (Strauss Lab) and Lindy Abas (Mach Lab)
"Salicylic Acid Targets Protein Phosphatase 2A to Attenuate Growth in Plants"
Summary
Plants, like other multicellular organisms, survive through a delicate balance between growth and defense against pathogens. Salicylic acid (SA) is a major defense signal in plants, and the perception mechanism as well as downstream signaling activating the immune response are known. Here, we identify a parallel SA signaling that mediates growth attenuation. SA directly binds to A subunits of protein phosphatase 2A (PP2A), inhibiting activity of this complex. Among PP2A targets, the PIN2 auxin transporter is hyperphosphorylated in response to SA, leading to changed activity of this important growth regulator. Accordingly, auxin transport and auxin-mediated root development, including growth, gravitropic response, and lateral root organogenesis, are inhibited. This study reveals how SA, besides activating immunity, concomitantly attenuates growth through crosstalk with the auxin distribution network. Further analysis of this dual role of SA and characterization of additional SA-regulated PP2A targets will provide further insights into mechanisms maintaining a balance between growth and defense.
» https://doi.org/10.1016/j.cub.2019.11.058
⇒ December 2019
Neues aus dem Internet der Pilze
Wenn ein Pilzgeflecht eine Nahrungsquelle gegen Bakterien verteidigt oder auf sonstige Art mit seiner Umwelt interagiert, dann sehen Forscherinnen und Forscher der Plattform BiMM (Bioactive Microbial Metabolites) in Tulln ganz genau hin. Das Wissen um die molekularen Prozesse und die "Links" im "Internet der Pilze" könnte sich sogar als Waffe gegen Krebs oder multiresistente Keime erweisen...
⇒ November 2019
BiRT Seminar Series „Die Welt der biologischen Interaktionen“
Dr. Sabine Fillinger
INRA-Institute National de la Rechérche Agronomique, AgroParisTech, Paris-Saclay, France
"Target & Non target-site resistance mechanisms against agricultural fungicides, the tip of the iceberg?"
Freitag 22. November 2019
10:00 Uhr
Campus Tulln / UFT – Seminarraum 16
Host: Joseph Strauss
⇒ October 2019
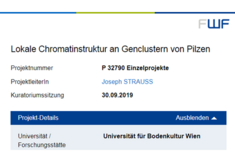
FWF Project for Joseph Strauss
"Chromatin composition at fungal gene clusters"
Project number: P 32790
Decision board: 2019/09/30
Keywords: filamentous fungi, chromatin, secondary metabolism, gene cluster regulation, Aspergillus
⇒ September 2019
Congratulations!
Congratulations to Anna Atanasoff-Kardjalieff who successfully defended her Master Thesis on Fusarium heterochromatin and epigenetics.
Very well done and welcome to our group as PhD student!
⇒ August 2019
Publication: "Evidence of a Demethylase-Independent Role for the H3K4-Specific Histone Demethylases in Aspergillus nidulans and Fusarium graminearum Secondary Metabolism"
Abstract
Fungi produce a plethora of secondary metabolites (SMs) involved in cellular protection, defense, and signaling. Like other metabolic processes, transcription of SM biosynthesis genes is tightly regulated to prevent an unnecessary use of resources. Genes involved in SM biosynthesis are usually physically linked, arranged in secondary metabolite gene clusters (SMGCs). Research over the last decades has shown that chromatin structure and posttranslational modifications (PTMs) of histones represent important layers of SMGC regulation. For instance, trimethylation of histone H3 lysine 4 (H3K4me3) is a PTM typically associated with promoter regions of actively transcribed genes. Previously, we have shown that the H3K4me3-specific, JmjC domain-containing histone demethylase KdmB functions not only in repression but also in activation of secondary metabolism in Aspergillus nidulans, suggesting that KdmB has additional functions apart from histone demethylation. In this study, we identified demethylase-independent functions of KdmB in transcriptional regulation of SM gene clusters. Furthermore, we show that this activating and demethylase-independent role of the H3K4 demethylase is also conserved in the phytopathogenic fungus Fusarium graminearum. Lack of FgKdm5 resulted in significant downregulation of five of seven analyzed SMs, whereby only one SMGC depends on a functional JmjC-domain. In A. nidulans strains deficient in H3K4 methylation, i.e., cclA∆, largely phenocopied kdmB∆, while this is not the case for most of the SMs analyzed in Fusarium spp. Notably, KdmB could not rescue the demethylase function in ∆fgkdm5 but restored all demethylase-independent phenotypes.
» https://doi.org/10.3389/fmicb.2019.01759
⇒ June 2019
Kinderuni am DAGZ
Am Montag und Dienstag (17./18.06.2019) waren je eine Klasse der Volksschule Familienschule Leopoldstadt bei uns zu Gast, jeweils für den ganzen Vormittag.
Die Kinder waren zwischen 8 und 10 Jahre alt (3. und 4. Klasse), es waren einmal 20 und einmal 21 Kinder, und wir hatten ein Programm mit Experimenten zum Thema Pflanzen und Pilze, und generell zur Arbeit in einem Biologie-Labor.
Die Mitwirkenden waren: Jeanette Moulinier-Anzola, Barbara Korbei, Elena Feraru, Sabine Strauss-Goller, Max Schwihla, Kai Dünser und Doris Lucyshyn.
» Korbei Lab
» Lucyshyn Lab
» Kleine-Vehn Lab
» Strauss Lab
Wissenschaftskommunikation "extended" - Wie beeinflussen Bakterien die Genexpression in assoziierten Pilzen?
Die eLIFE Publikation über Bakterien-Pilz Interaktion der Arbeitsgruppe Strauss, wurde als Highlight in der Märzausgabe der Zeitschrift Biospektrum vorgestellt.
» https://doi.org/10.1007/s12268-019-1048-4 (Biospektrum)
» https://www.ncbi.nlm.nih.gov/pubmed/30311911 (eLIFE)
⇒ May 2019
Publication: "Fusaoctaxin A, an Example of a Two-Step Mechanism for Non-Ribosomal Peptide Assembly and Maturation in Fungi"
Abstract:
Fungal non-ribosomal peptide synthetase (NRPS) clusters are spread across the chromosomes, where several modifying enzyme-encoding genes typically flank one NRPS. However, a recent study showed that the octapeptide fusaoctaxin A is tandemly synthesized by two NRPSs in Fusarium graminearum. Here, we illuminate parts of the biosynthetic route of fusaoctaxin A, which is cleaved into the tripeptide fusatrixin A and the pentapeptide fusapentaxin A during transport by a cluster-specific ABC transporter with peptidase activity. Further, we deleted the histone H3K27 methyltransferase kmt6, which induced the production of fusaoctaxin A.
» https://doi.org/10.3390/toxins11050277
⇒ March 2019
⇒ February 2019
DAGZ Tulln Seminar
"Key findings on how different nitrogen sources affect root development in Arabidopsis thaliana" Great insights how different forms of nitrogen nutrition influence plant root development in the yeasterday DAGZ Tulln seminar given by IST researcher Krisztina Ötvös. » https://www.researchgate.net/profile/Krisztina_Oetvoes
Regulation of a novel Fusarium cytokinin in Fusarium pseudograminearum
Abstract
Fusarium pseudograminearum is an agronomically important fungus, which infects many crop plants, including wheat, where it causes Fusarium crown rot. Like many other fungi, the Fusarium genus produces a wide range of secondary metabolites of which only few have been characterized. Recently a novel gene cluster was discovered in F. pseudograminearum, which encodes production of cytokinin-like metabolites collectively named Fusarium cytokinins. They are structurally similar to plant cytokinins and can activate cytokinin signalling in vitro and in planta. Here, the regulation of Fusarium cytokinin production was analysed in vitro. This revealed that, similar to deoxynivalenol (DON) production in Fusarium graminearum, cytokinin production can be induced in vitro by specific nitrogen sources in a pH-dependent manner. DON production was also induced in both F. graminearum and F. pseudograminearum in cytokinin-inducing conditions. In addition, microscopic analyses of wheat seedlings infected with a F. pseudograminearum cytokinin reporter strain showed that the fungus specifically induces its cytokinin production in hyphae, which are in close association with the plant, suggestive of a function of Fusarium cytokinins during infection. » https://doi.org/10.1016/j.funbio.2018.12.009
⇒ January 2019
"A novel laminar-flow-based bioaerosol test system to determine biological sampling efficiencies of bioaerosol samplers"
Abstract
In this work, we describe a novel type of bioaerosol test system based on a laminar airflow chamber that provides a homogenous aerosol of microbial cells with known concentrations and defined culturability to bioaerosol samplers positioned in the chamber. In the system, three control and monitoring points (CMPs) are implemented in which the number and culturability of microbes can be determined by combining optical particle counting with microscopic and culture-based microbiological analyses... » https://doi.org/10.1080/02786826.2018.1562151
Paper from BiMM with the AKH
"Antifungal susceptibility of yeast blood stream isolates collected during a 10 year period in Austria" Background
Candida‐associated infections put a significant burden on western health care systems. Development of (multi‐) resistant fungi can become untreatable and threaten especially vulnerable target groups, such as the immunocompromised. Objectives
We assessed antifungal susceptibility and explored possible influence factors of clinical Candida isolates collected from Austrian hospitals between 2007 and 2016 » https://doi.org/10.1111/myc.12892 » BiMM Bioactive Microbial Metabolites
⇒ December 2018
Publication from Lena Studt
"The putative H3K36 demethylase BcKDM1 affects virulence, stress responses and photomorphogenesis in Botrytis cinerea" Highlights:
- T-DNA insertion in the virulence-attenuated mutant PA2810 disrupts bckdm1.
- Bckdm1 encodes a putative histone 3 lysine 36-specific demethylase.
- Bckdm1 is required for virulence, stress responses and photomorphogenesis.
- Orthologs from other Ascomycetes cannot replace BcKDM1.
» https://doi.org/10.1016/j.fgb.2018.11.003
⇒ November 2018
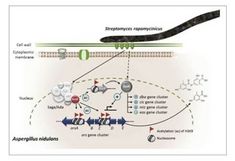
"Chromatin mapping identifies BasR, a key regulator of bacteria-triggered production of fungal secondary metabolites"
Fungi fight back with epigenetic mechanisms when bacteria steal their nutrients.
How it works is elucidated in a new paper from the Strauss group in collaboration with the Leibniz-Institute for Infection Biology Jena. see details on https://www.ncbi.nlm.nih.gov/pubmed/30311911 The eukaryotic epigenetic machinery can be modified by bacteria to reprogram the response of eukaryotes during their interaction with microorganisms. We discovered that the bacterium Streptomyces rapamycinicus triggered increased chromatin acetylation and thus activation of the silent secondary metabolism ors gene cluster in the fungus Aspergillus nidulans...
BiRT Lecture Series
Dr. Jan-Peter George
BFW Bundesforschungszentrum für Wald
Institut für Waldgenetik "Molecular genetics reveal putative adaptive variation in resistance against Chrysomyxa rhododendri in Norway spruce" Thursday November 22nd, 2018
Time: 10:00
Campus Tulln / UFT – Seminar Room 14
Host: Joseph Strauss & Markus Gorfer
⇒ October 2018
Die BiMM und das Internet der Pilze
Auf der Suche nach Links im Internet der Pilze
Können Schimmelpilze Informationen über die Abwehr von Bakterien an das gesamte Pilznetzwerk weitergeben und speichern?
Die Antwort auf diese Frage könnte bei der Entwicklung neuer Antibiotika und Krebstherapien helfen » zum Artikel BiMM Bioactive Microbial Metabolites
⇒ September 2018
Fungus found floating in the Danube river - now identified and described by BiMM team as new fungal species
"Metapochonia lutea, a new species isolated from the Danube river in Austria" A new species Metapochonia lutea (Ascomycota, Hypocreales) is described and illustrated. This fungus differs from the other taxa in the genus Metapochonia by its production of intensive yellow pigment in culture, conidiophores with relatively complex verticillate branching, bean-shaped conidia, and by delayed development of one-celled and prominent thick-walled submerged chlamydospores usually in chains or irregular clusters of 3–5 cells... https://www.schweizerbart.de/journals/nova_hedwigia
WKO Preis 2018 für Andreas Schüller
Andreas Schüller stellt sein Projekt vor, welches mit dem Wirtschaftskammerpreis 2018 an der BOKU ausgezeichnet wurde. Es geht um die Etablierung eines neuen Systems zur gezielten Aktivierung und Entdeckung von bioaktiven Stoffen in filamentösen Pilzen. https://www.youtube.com/watch?v=3BCayifmbxo
⇒ August 2018
Just cut it - an easy way to turn a negative regulator to positive mode-of-action
"Truncation of the transcriptional repressor protein Cre1 in Trichoderma reesei Rut-C30 turns it into an activator" Background
The filamentous fungus Trichoderma reesei is a natural producer of cellulolytic and xylanolytic enzymes and is therefore industrially used. Many industries require high amounts of enzymes, in particular cellulases. Strain improvement strategies by random mutagenesis yielded the industrial ancestor strain Rut-C30... » https://doi.org/10.1186/s40694-018-0059-0
⇒ July 2018
FWF Project for Lena Studt - Congratulations!
"Role of the histone variant H2A.Z in phytopathogenic fusaria"
project number: I 3911 Internationale Projekte decision board: 25.06.2018 » Lena Studt
Publication from Lena Studt
"Set1 and Kdm5 are antagonists for H3K4 methylation and regulators of the major conidiation‐specific transcription factor gene ABA1 in Fusarium fujikuroi" Summary
Here we present the identification and characterisation of the H3K4‐specific histone methyltransferase Set1 and its counterpart, the Jumonji C demethylase Kdm5, in the rice pathogen Fusarium fujikuroi. While Set1 is responsible for all detectable H3K4me2/me3 in this fungus, Kdm5 antagonises the H3K4me3 mark. Notably, deletion of both SET1 and KDM5 mainly resulted in the upregulation of genome‐wide transcription, also affecting a large set of secondary metabolite (SM) key genes... » https://doi.org/10.1111/1462-2920.14339
⇒ June 2018
Agrargenetik und Biodiversität - Exkursion 20.06.2018
Das war auch heuer wieder ein spannender Exkursionstag im Zoo Schönbrunn mit Vorträgen und Backstage-Führungen für unsere Studierenden des Bachelorstudiums Agrarwissenschaften.
Die Themen: Warum ist Biodiversität nur mit genetischer Diversität denkbar, warum ist Artenschutz essentiell und was sind die Gründe für die „Kernschmelze der Agrar-Biodiversität“ in den letzten Jahrzehnten?
Diese und viele weitere Fragen wurden von den Studierenden mit den Veranstaltern diskutiert. Ein Angebot des DAGZ im Zuge der Lehrveranstaltung "941091 Agrargenetik und Biodiversität - Exkursion".
Gesichter der BOKU - David Cánovas
Interview mit David Cánovas Mitarbeiter Fungal Genomics Unit - Universitäts- und Forschungszentrum Tulln https://youtu.be/iSoieigVGv0
⇒ May 2018
New ERASMUS student Safa Oufensou arrived to our lab from Sardinia.
Wheather in Tulln will not be as good as on her home island, but sure she will have a great time in our lab. Welcome Safa!
⇒ January 2018
⇒ December 2017
Is cell-wall stress sensed for secondary metabolite gene expression?
A first insight into this possible connection was published this summer in collaboration with Gea (Guerriero), former Lise-Meitner Grant fellow in our lab and now group leader at Luxemburg Institute of Science and Technology.
…and at the same time warm welcome to our two new lab members
# Franz (Zehetbauer), who started his PhD on the epigenetics of cellular communication, and
# Astradeni, our ERASMUS guest student from the Diallinas Lab in Athens/Greece
⇒ August 2017: Former Bac student Tamara famous in Austrian media
Tamara, a former Bac-student of the group now at Kansas University and already featured in Austrian press as young researcher in fungal genetics »read full article.
⇒ June 2017: New publication in "Scientific Report"
The paper describes how to use robots and check how quickly fungi grow in their natural solid substances"
⇒ June 2017: Exkursion "Agrargenetik & Biodiversität" in den Zoo Schönbrunn
Das unbestrittene Highlight des Jahres was unsere Lehre betrifft: die Exkursion "Agrargenetik & Biodiversität" in den Zoo Schönbrunn.
Danke an das Zoo-Team um Wiebke Hoffmann!
⇒ June 2017: New "collaborative-PhD" Alice Rassinger
⇒ April 2017: Symbiocyte - Start-Up eines BOKU-DAGZ Wissenschaftlers gewinnt Innovationspreis GENIUS 2017
Symbiocyte entwickelt innovative Biopestizide und konnte sich damit im diesjährigen GENIUS Innovationspreis des Landes Niederösterreich gegen zahlreiche Bewerber durchsetzen und den ersten Platz erreichen.
Das Unternehmen wurde 2016 von Dr. Harald Berger gegründet, ein Spezialist für funktionelle Genomforschung in Pilzen und langjähriger Mitarbeiter in der Pilzgenetik Forschungsgruppe von Prof. Joseph Strauss am Department für Angewandte Genetik in Tulln ...⇒ more information
Kontakt: Dr. Harald Berger harald.berger(at)symbiocyte.com - 0664/5366690
Dr. Alexander Pretsch alexander.pretsch(at)symbiocyte.com - 0664/2443774 Symbiocyte
Technopark 1 / Geb C / EG
3430 Tulln
⇒ April 2017: Farewell to Lucia
After several years at BOKU in our lab and at Department IFA Tulln (group Ines Fritz) Lucia left us - but for a nice reason: she took up a position at the Yeast laboratory of Prof. Bernardi to go ahead with what she initially started to study and came to us in 2009 for a “short stay” to learn in vivo footprinting and ChIP. In the end the “short stay” lasted seven very productive and pleasant years.
Lucia, we´ll miss you but looking forward to nice collaborations!
Dr. Lucia Silvestrini
Yeast Laboratory (Prof. Berardi)
Università Politecnica delle Marche
Department of Agricultural, Food and Environmental Sciences
Ancona, Italy